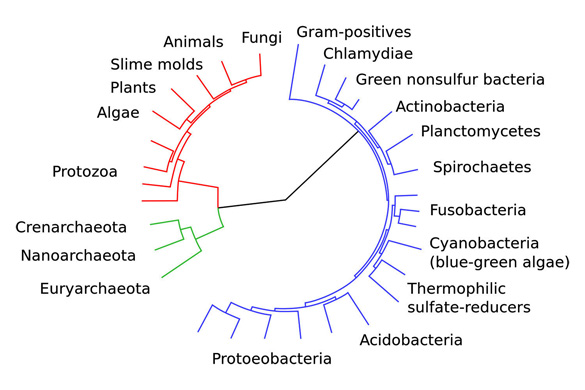
 This image is of an evolutionary tree—a so-called ‘Tree of Life’—showing the divergence of modern species from their common ancestor in the center. The three domains are coloured, with bacteria blue, archaea green and eukaryotes red. Courtesy/Wikipedia Commons
This image is of an evolutionary tree—a so-called ‘Tree of Life’—showing the divergence of modern species from their common ancestor in the center. The three domains are coloured, with bacteria blue, archaea green and eukaryotes red. Courtesy/Wikipedia Commons
LANL News:
- New technology puts bioinformatics within easy reach of health-care professionals, researchers and others
Los Alamos National Laboratory has released an updated version of powerful, award-winning bioinformatics software that is now capable of identifying DNA from viruses and all parts of the Tree of Life—putting diverse problems such as identifying pathogen-caused diseases, selection of therapeutic targets for cancer treatment, and optimizing yields of algae farms within relatively easy reach for health-care professionals, researchers and others.
“As part of our testing, we used Sequedex to identify virus sequences in a collaborator’s clinical blood sample from Africa,” said Ben Mcmahon, a scientist in Los Alamos’s Theoretical Biology and Biophysics group. “In the course of an afternoon, the software had identified a deadly rabies virus, something that would have taken weeks of work using conventional methods. Sequedex software can now identify sequences from viruses and fungi at parts-per-million levels in a sequenced sample.”
The new Version 1 edition of Sequedex recognizes patterns in short DNA sequences, and then associates those sequences with phylogeny—the sample’s placement on the evolutionary Tree of Life—and the function of the fragment. In evolutionary terms, a “Tree of Life” is a representation of the genetic divergence of modern species from a common ancestor. Based on the recognition of the DNA pattern, the software creates a database of results.
Sequedex classifies fragments 250,000 times faster than conventional methods. With Sequedex, a laptop computer can analyze DNA sequences faster than any current DNA sequencer can create them.
Los Alamos researchers designed the software to perform bioinformatics without the need for a bioinformatician to perform calculations and interpret the results.
Sequedex analyzes phylogeny and function in a collection of DNA sequences in a similar fashion to doing a search in a web browser. For example in Google, entering the search terms “plumber”, “Smith”, and “Chicago” might return links to plumbers named Smith in the Windy City; similarly, Sequedex uses a list of search terms generated from previously classified genomes to link phylogeny and function to DNA sequences. The search terms generated by Sequedex are selected by evolution in the sense that they must be present in more than one genome. Each term is also linked to a branch of the Tree of Life and a set of one or more biological functions.
As an example, in a code that is one letter per amino acid, the protein pattern “CVELAHEIRS” is found in humans and mice, so Sequedex associates it with the phylogenetic classification Chordates, to which both humans and mice belong. In humans, CVELAHEIRS is found in a protein classified as a “Regulator of G-protein Signaling” (or RGS for short), so Sequedex also associates the term with the RGS function. When Sequedex finds CVEHLAHEIRS in a DNA sequence (translated into protein sequences via the genetic code), it identifies the sequence as likely coming from a Chordate RGS.
The chance of finding CVELAHEIRS in a stretch of DNA by random chance is low, so even when the search term comes from an organism that Sequedex doesn’t know about (for example, yaks, killer whales, and naked mole rats are not currently in the Sequedex Library but all have CVELAHEIRS in their genomes) the software still has a good chance of making the correct family and functional identification.
Sequedex holds promise for use in identifying infectious diseases in clinical samples; characterizing the spaces within the human body that are shared by other organisms, and how these so-called microbiomes are associated with health or disease; and analyzing tumor genetics for chemotherapy options and prognosis. Other features of Sequedex V1 include the ability to self-update and make plots of results. The software, however, is applicable right now only as a research tool; it is not intended to diagnose a disease or other condition.
Los Alamos scientists Benjamin McMahon, Nick Hengartner, Judith Cohn, Mira Dimitrijevic, and Joel Berendzen developed Sequedex.
The breakthrough technology received a 2012 R&D 100 award, earning distinction as one of R&D Magazine’s 100 most-significant inventions of the year. The software has been beta tested for the past two years by nearly 50 labs around the world. Sequedex is available for licensing. To learn more visit LANL’s Richard P. Feynman Center for Innovation website here or contact the Licensing Team at the FCI at licensing@lanl.gov.
Sequedex V1 is available under a free six-month demonstration license. It may be downloaded from http://sequedex.lanl.gov
About Los Alamos National Laboratory
Los Alamos National Laboratory, a multidisciplinary research institution engaged in strategic science on behalf of national security, is operated by Los Alamos National Security, LLC, a team composed of Bechtel National, the University of California, The Babcock & Wilcox Company, and URS Corporation for the Department of Energy’s National Nuclear Security Administration.
Los Alamos enhances national security by ensuring the safety and reliability of the U.S. nuclear stockpile, developing technologies to reduce threats from weapons of mass destruction, and solving problems related to energy, environment, infrastructure, health, and global security concerns.

